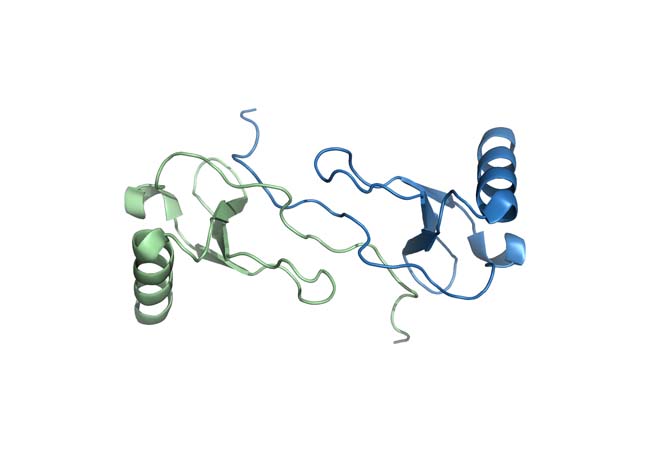

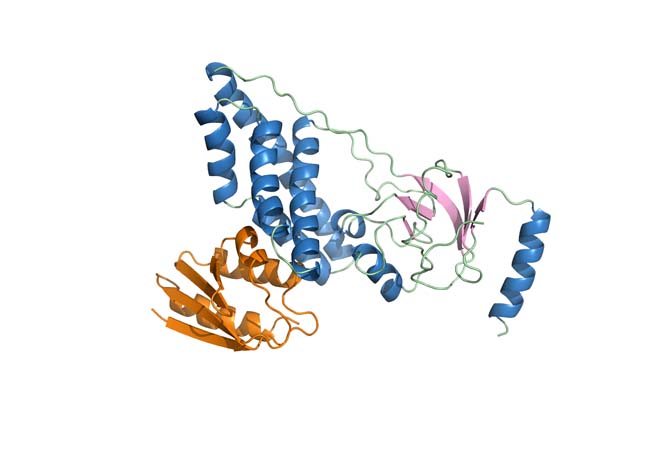
Traditionally, the determination of three-dimensional structures of biological macromolecules by NMR has employed distance and torsion angle constraints. Despite its enormous successes, the strictly local nature of the experimental constraints can pose undesirable limitations to the attainable accuracy, especially with respect to the relative orientation of individual structural elements. This shortcoming was overcome by the development of that exploits weak alignment of molecules in the magnetic field which permits the determination of orientational constraints from residual dipolar couplings (RDC). We are using paramagnetic relaxation enhancements (PRE) to extract supplemental restraints in cases where NOE data is found limiting, especially in multicomponent complexes.
19F NMR
While naturally occurring magnetically active isotopes, such as 1H, 13C,15N, are most commonly used in biomolecular NMR, with 15N and 13C isotopic labeling routinely employed at the present time, 19F is a very attractive and sensitive nucleus, which offers rich information on molecules in solution and in the solid state. The overall goal of our research program on 19F NMR is to establish a versatile and ubiquitous approach for elucidating structure and dynamics in proteins, in solution and in the solid state.
In particular, we developed 19F PRE-based methods for extracting quantitative distances. The largest advantage of using 19F PRE-based distance measurements over proton PREs is the fact that spectral overlap does not impede their extraction and that any specific distances can be targeted by judicious placement of the paramagnetic label as well as the fluorine atom(s). This is especially beneficial in the study of large proteins, protein complexes and membrane proteins.
Other specific objectives are to utilize 19F chemical shifts as a probe of geometry, electronic structure, chemical reactivity, and dynamics in a variety of biologically important molecules through the integration of experimental 19F solution and solid-state NMR parameters with quantum chemical calculations. Given the simplicity of biosynthetic incorporation of fluorine into a wide category of specific and unique sites and the very large 19F chemical shift range this approach has the potential to transform the field.